Artificial intelligence predicts the shape of human proteins and opens a new era in biology - the counter
Scientists announced in Nature magazine the availability of the largest protein database that form the structures of life, which "will fundamentally change research in biology," according to specialists.
Fundamental pieces of life, the structure of each protein, which depends on the amino acids that compose it, define what it does and how it does, so that determine it provides valuable information to understand biological processes, advance various fields to investigate andcould serve for drug development.
Google Deepmind
The DEEPMIND company and the European Laboratory of Molecular Biology (EMBL) have used the Alphafold artificial intelligence system to publish the most complete and precise database of the predictions of the structures of human proteins.
Each cell of a living organism executes its function with the help of proteins that permanently instructions to keep the cell in good health and combat infections.Unlike genome - the sequence of genes that codify cell life - the human proteoming permanently changes in response to genetic instructions and exterior stimuli.
Understanding the functioning of proteins, through the way they adopt inside the cells, is a real challenge.
Only 17 % of the human proteom components are known
Scientists have applied to determine their precise function through experiments.But after 50 years of research, only 17 % of amino acids, or human proteom components are known.
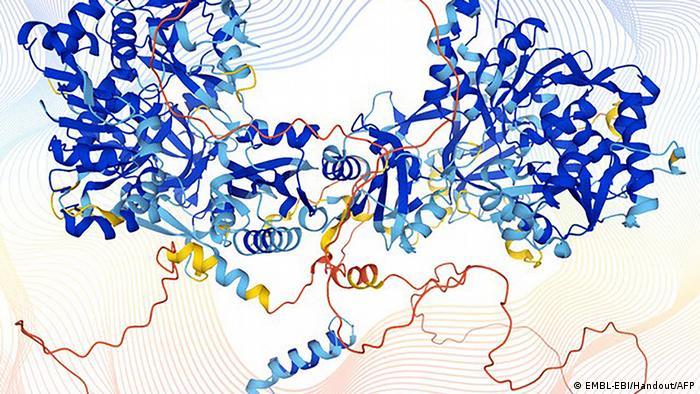
Google Deepmind researchers and the European Molecular Biology Laboratory (EMBL) revealed on Thursday a free access database, 20.000 proteins manifested by the human genome.To which 350 are added.000 proteins of 20 organisms, such as bacteria or mice, used for research.
How to boot from usb drive to install windows, ubuntu, or Osx on surface pro devices - https: // t.co/qnf4or3qor https: // t.CO/1Oys6mvidf
— alpinetworkcom Fri Apr 21 22:05:57 +0000 2017
Alphafold: Automatic Learning Program
This base was obtained thanks to an automatic learning program capable of precisely predicting the shape of a protein from its amino acid sequence.
The Alphafold program trained with 170.000 known protein structures and then predicted the form of 58 % of all human protein proteins, which most doubled the number of human protein structures known precisely.
Of these, the position of a subset of 35.7 % was predicted with a "very high" degree of trust, which is twice the number covered by experimental structures, explained the magazine.
Large -scale structures prediction
The researchers consider that the prediction of large -scale and precise structures will become "an important tool that will allow you to address new scientific issues from a structural perspective", and Alphafold's predictions will help to clarify even more the role of proteins.
"We believe that this is the most significant contribution that artificial intelligence has done to the progress of scientific knowledge to date, and is a great example of the types of benefits that artificial intelligence can contribute to society," according to the founder of Deepmind, Demis Hassabis, British firm that belongs to Alphabet, Google Matrix.
The use of artificial intelligence, with its ability to computationally predict the shape of a protein from its amino acid sequence, allows you not to have to determine experimentally with the use of laborious and sometimes expensive techniques.
GENETIC DISEASE RESEARCH
The potential applications of these data go from research on genetic diseases to drought -resistant harvest engineering.
According to Paul Nurse, Nobel Prize in Medicine and director of the Francis Crick Institute, this advance is "a great step for biology innovation".
John McGeehan, director of the Enzyme Innovation Center at the University of Portsmouth, stressed that "what he took months and years to be fulfilled was carried out in a weekend by Alphafold".
The ability to predict with a computer program the shape of a protein from its sequence of loved acids is already being applied in some sectors of the investigation.
